I needed a break from the neural network and even machine learning algorithms. All those formulae for mathematical representation, the complexity of all those algorithms were getting on my nerves.
So, today I was looking for something that is fresh and fun. Look what I found! It’s RSQLite.
What is RSQLite?
RSQLite embeds the SQLite database engine in R.
So, What is SQLite?
SQLite is an embedded SQL database engine. It is a public-domain, single-user, very light-weight database engine that implements a decent subset of the SQL 92 standard, including the core table creation, updating, insertion, and selection operations, plus transaction management. SQLite does not have a separate server to process.
It reads and writes directly to ordinary disk files. A complete SQL database with multiple tables, indices, triggers, and views, is contained in a single disk file.
Why RSQLite?
If the size of a dataset is too large to handle and for the analysis, only a few features can be considered at a time or we need a chunk of the data for analysis, then storing the data outside of R and organizing it in a database can be a good idea.
Many large datasets are already available in public or private databases. So, we can simply query them without having to download the data first.
How to install SQLite in R?
The latest released version of RSQLite can be installed from CRAN with the below command:
install.packages("RSQLite")
How to connect to SQLite database in R?
First, Load the “RSQLite” library.
library("RSQLite")
Then, set R’s working directory to the directory where we would like to create the database.
# Set working directory
setwd("/Users/oindrilasen/WORK_AREA/Data Science/Sqlite")
Now, we can connect R to the SQLite database file using dbconnect() and will create a temporary in-memory database.
# Create an in-memory RSQLite database con = dbConnect(SQLite(), ":memory:") con
How to work on the SQLite database in R?
Now, let’s work on some basic operation on the SQLite Database:
dbWriteTable(con, "mtcars", mtcars)
alltables = dbListTables(con) alltables
dbListFields(con, "mtcars")
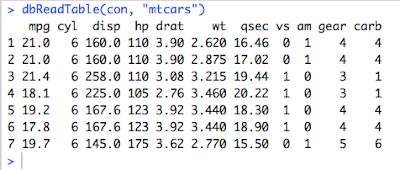
4. Read the Table created above.
dbReadTable(con, "mtcars")
5. Build a query (i.e., describe what we want to fetch from the SQLite database). In the query below, we will be grabbing all the columns from the table named “mtcars” from the database we just connected to where cyl value is 6.
myQuery <- dbSendQuery(con, "SELECT * FROM mtcars where cyl = 6")
6. After building the query, we can use that query to fetch the data from the database and assign that to an R object. The n = -1 argument in dbFetch() must be used if we want to retrieve all records in the database, otherwise, the default is to stop at 500 records.
mtcars <- dbFetch(myQuery, n = -1) str(mtcars)
7. After assigning the data to an object in R, we need to clear the results.
dbClearResult(myQuery)
And, that’s all for today!